A search to Medications to treat Covid-19 via Computational Chemistry methods and Molecular Docking
In December 2019 the first cases of infection from a novel coronavirus (Covid-19) were reported. Since then Covid-19 is spreading at an alarming rate and has created an unprecedented health emergency around the globe. The virus has infected more than 3,000,000 people and 217,000 have died.
There is no effective vaccine and it will most likely take at least 1-1.5 year to develop one. Therefore the development of antiviral agents is an urgent priority even though it usually takes many years for new drugs to be discoverd, clinically tested and approved. A good strategy would be trying to find already approved drugs that have some efficacy against similar type of viruses. Then test the efficacy of these drugs using computational chemistry methods and molecular docking. The most effective of these drugs can then be clinically tested and approved. There is a great deal of open access data available on the Internet.
It should be mentioned that computational chemistry methods and molecular docking results of drug efficacies not to be taken as medical advice or evidence supporting a specific treatment.
The steps required for drug screening are as follows:
- Find the 3D molecular structure of Covid-19 from the >Protein Data Bank PDB.
- Find the molecular structures of drugs (ligands) with efficacy against similar viruses.
- Find the ground state optimization of these drugs (ligands).
- Calculate the HOMO - LUMO gap (energy difference gap between the HOMO and LUMO molecular orbitals) at the ground state molecular geometry of the drug (ligand) (from the above step).
- The interaction of the above mentioned drugs (ligands) with Covid-19 5R82 protease (receptor) were determined using Molecular Docking.
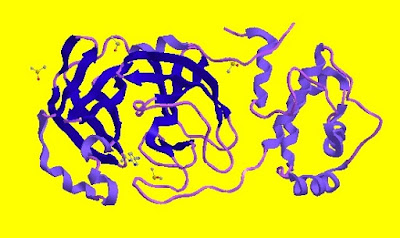
The crystallized main protease of Covid-19 (5R82) can be selected. It is shown in Fig. I.1 below:
Hundrends of drugs can be found. A great deal of open access data are available on the Internet. Some of them are: remdesivir, chloroquine, colchicine, darunavir, favipavir, oceltamivir, niclosamide. The 3D and 2D structures of remdesivir is shown in Fig. I.2 below:
The ground state optimization of a compound is the molecular geometry with the lowest energy (the most stable molecular geometry). There are several computational chemistry softwares that use semiempirical and ab initio methods to obtain the ground state geometry of a compound. Some of them are: Gamess, Gaussian, Orca, Avogadro, Firefly, Arguslab. The Arguslab software was used and the PM3 method was selected. This method is semiempirical and fast but not as accurate as the high level ab intio methods. PM3, or Parametric Method 3, is based on the Neglect of Differential Diatomic Overlap integral approximation. The ground state optimized structure of favipavir using the PM3 method is shown in Fig. I.3:
The energy gap between the HOMO (highest occupied molecular orbital) and the LUMO (lowest unoccupied molecular orbital) is an important quantum chemical parameter that characterizes the chemical reactivity of a molecule. A molecule with a small energy gap is more reactive compared to a molecule with a large HOMO - LUMO gap. The HOMO - LUMO gap energy of each drug was calculated and is shown in Table I.1
The interaction of drugs with Covid-19 5R82 protease (receptor) were determined using two softwares Autodock Vina and iGemDock. During the docking process, the receptor and the ligand are rotated around their own coordinate origin and the separation between the two origins is varied. A score is calculated for each orientation and the lower binding energy obtained corresponds to the best interaction between the receptor and the ligand (highest binding affinity).
The results obtained are shown in Table I.1 below:
| Table I1: Quantum Chemical & Docking Results for the Interaction of selected Drugs with Covid-19 5R82 protease | ||||||
|---|---|---|---|---|---|---|
| Compound | HOMO-LUMO energy gap (a.u.) | Docking iGemDock | Docking AutoDock - Vina (kcal/mol) | |||
remdesivir |
0.299555 |
|
|
|||
chloroquine |
0.292904 |
|
|
|||
hydroxychloroquine |
0.292930 |
|
-5.9 |
|||
colchicine |
0.305204 |
|
-6.4 |
|||
darunavir |
0.315079 |
|
-7.7 |
|||
favipavir |
0.328258 |
|
-5.8 |
|||
oceltamivir |
0.343420 |
|
-5.7 |
|||
As it can be seen from Table I.1 the lowest HOMO-LUMO gaps at the PM3 level are observed for chloroquine, hydroxychloroquine and remdesivir. These molecules are the most reactive. However, the docking results for these three drugs show that remdesivir has the lowest binding energy and therefore the highest binding affinity for the receptor.
Amongst all the drugs listed in Table I.1 the highest binding affinity for the protease of Covid-19 5R82 is shown by darunavir (-7.7 kcal/mol) according to Autodock Vina (the second highest affinity is shown by colchicine -6.4 and the third by remdesivir -6.3 kcal/mol) . Darunavir shows the second highest affinity according to iGemDock (-141.5402) while remdesivir scores a bit higher (-144.26573).
It is worth stressing that binding is not synonymous with inhibition. Even the most well bound molecule may have little effect on a protein if it targets the wrong site. One limitation of the work is the choice of binding site all the above drugs were tested against. This was constrained by the ligand used to stabilize the protein crystal structure, RZS. If DMS was used as the ligand instead of RZS, the drugs would have been tested against different sites and different results may have found.
For a similar post using a SARS-CoV-2 spike protein as a receptor see the link below.
Relevant Posts - Relevant Videos
References
- M. A. Thompson, “Molecular docking using ArgusLab, an efficient shape-based search algorithm and AScore scoring function,” in Proceedings of the ACS Meeting, Philadelphia, Pa, USA, March-April 2004, 172, CINF 42.
- O. Trott, A. J. Olson, "AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading", Journal of Computational Chemistry 31 (2010) 455-461
- Hsu, K., Chen, Y., Lin, S. et al. iGEMDOCK: a graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinformatics 12, S33 (2011).
Key Terms
covid-19,HOMO , LUMO,ground state optimization, medication for covid-19, drugs for covid-19, molecular docking, ab initio methods, computational chemistry, binding energy, binding affinity,

